Generating Syntethic Data for Causal-Inference
The program for generating synthetic data is located in - H:\MR\Projects\Shared\CausalEffects\CausalEffectsUtils\generate_realistic_data
the program implements the following model -
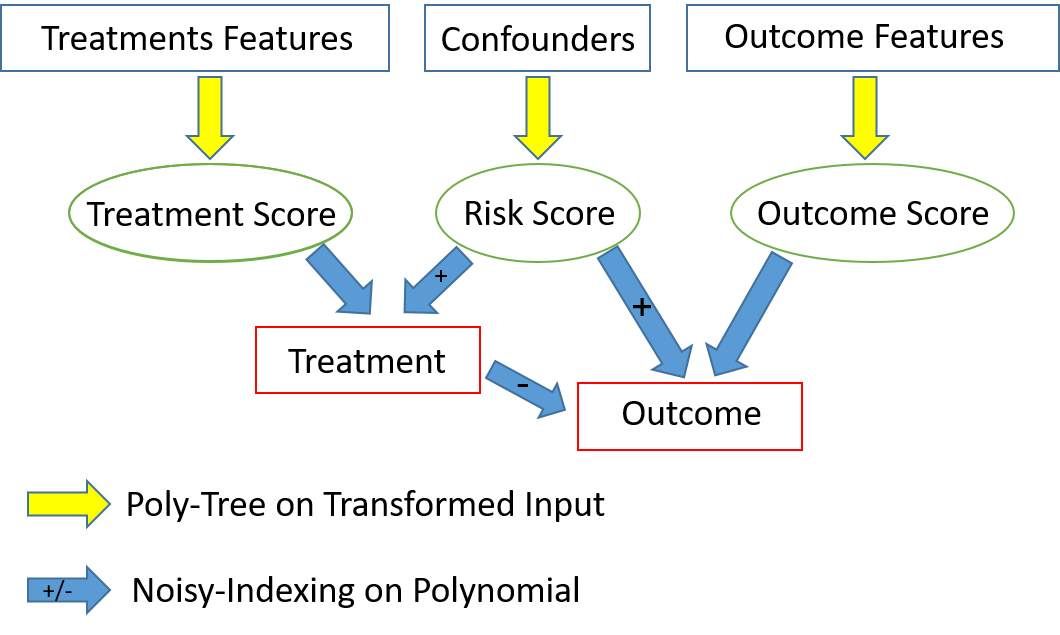 Where -
Where -
- Poly-Tree = Tree with polynomials at the nodes
- Transformed-input = apply the following transformation before calculating the polynomial (currently n=3 is hard-coded):
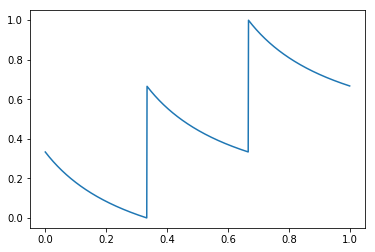
- Noisy-Indexing = apply logistic function on value, with minimal/maximal values moved from [0.0,1.0] to [ε,1-ε'], and then use that as probability for deciding on the dichotomic Treatment/Outcome
- +/- = requiring that the first-order contributions of a parameter to the polynomial is set to be positive (negative)
-
Parameters for running the generation are:
-
Note that -
- Currently, the depth of the trees is hard-coded as depth=2
- SigWidth determine the scaling of the logistic function, normalized by the distribution of the input variable. The lower it is, the lower the slope at the step is (i.e., very large widths correspond to step function)
- Features are generated as Uniform[0,**]
- Treatment is scaled by treatmentFactor before application of final polynomial
- risk is a debugging output matrix giving various intermediate values (e.g. Outcome/Treatment/Risk scores, various probabilities, etc.)
- The models are written into *.bin and *.treatment.bin
- Additional projects in the same solution include -
- Generate matrices given the model:
The program generates a random feaures matrix (Uniform[0,]), either uses .treatment.bin to set the treatment or randomly selects it (if ** is given) for a Randomized Controlled Trial scenario, and then uses **.bin to set the output
- Various utilities for handling the synthetic data/model -
- Generate matrices given the model:
Possible modes include:
- print - get a human-readable version of a generative model (from params.bin****
- getProbs - generate a vector of the output probabilities given a matrix and a model
- getAUC - get the maximal possible AUC for outcome prediction (known true probabilities)
- csv2bin - translate a csv matrix to binary format (serialized MedFeatures)