Missing Values Based Uncertainty Analysis
Use case: KP, Lung Cancer, monthly samples (monthly, without suspected, at least 3 months before outcome for cases) Question / goal / idea: When we have missing values we can:
- a) ignore the patient (if he has 'too much missing info')
- b) impute the missing values, or
- c) using GIBBS estimation of score std to decide to on which patients it is more important to fill the missing values What is better? Setting: MR/Projects/Shared/AlgoMarkers/LungCancer/scripts_uncertainty/adjust_and_calculate.sh MR/Projects/Shared/AlgoMarkers/LungCancer/configs_uncertainty/GIBBS.json
Evaluation: Easier to start with a notebook, but we intend to add a function to bootstrap_app, to report result by Top N Options:
- a) One sample per patient (if you using bootstrap_app - we should chose the sample before the app)
- b) all samples per patient, or
-
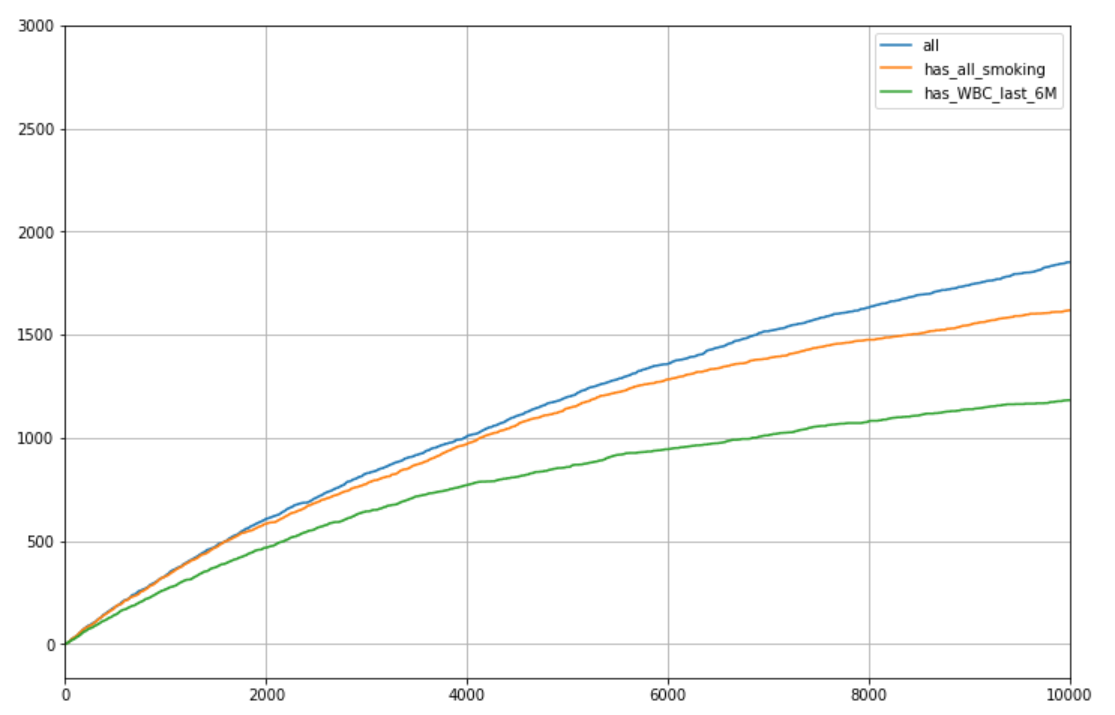
c) simulation (relatively complicated in this use case - hopefully we won't need it) Baseline Basic evaluation - in the graph, number of cases in the Top N, one sample for patient, age 50-80
-
if we take all patients (3215 on average)
- if we take just patients with smoking intensity information (1964 patients eligible on average)
- if we take just patients with WBC in the last 6 months (1438 eligible on average)
 A few notes:
A few notes: - Results are for Ex or Current smokers - if we included never smokers results would have been a bit better ...
-
I got similar results when we take all samples per patient Thus, it is better to take all patients, using the standard imputer, but we will stick to Ex or Current smokers Next, we will explore ways to be do even better using the variability in potential score, using GIBBS imputation statistics GIBBS - naïve Preliminaries:
-
The script adjust_and_calculate.sh train the adjusted model on all samples - DONE (leakage! should be re-visited later)
- However, apply (calculate) on all samples is far too slow
- So the Notebook: KP_uncertainty_generate_samples take one sample per patient, and apply GIBBS just for the sample
-
We now have 15 examples so we can test results variance Naive approach would be to replace pred_0 with pred_0 + score std based on GIBBS, to prioritize high uncertainty (or by pred_0 - score std for the opposite) Both options do not help. Furthermore, if we take (for instance) pred_0 +/- 3 X score std result deteriorates. Missing data completion Assume we have M tickets for data completion. We will choose the highest M patients with missing information, with the highest pred_0 + score std based on GIBBS.
-
For now we assume 100% compliance
- Note that it night not be optimal to take the highest M, as (when we not the working point and expected cutoff) those in the top with small std do not need data completion. However, they are not many, and as the results are not sensitive to M, we keep it this way for code simplicity, and to get results for several working points in one run. To simulate data completion:
- Optional, use data from the future:
- Relevant only for smoking features
- Data exists in ~1/3 of cases and ~1/3 of controls
- 2nd option, find the m (for instance m=20) closest patients
- Closets means same gender, same smoking status, distance is measured over all other (normalized) features ,with higher weight for age, and smoking features
- Choose 1 patient randomly from the 20, and take his values
- For patients with missing data but not in the top M, use imputation by our standard imputer (i.e., use original score)
- Implemented in Notebook: KP_uncertainty_missing_values_completion After that we re-calculate the score for those M patients (prediction is made based on split with the right model from the cross validation), take Top N and check performance. MAJOR DRAWBACK In smoking features calculation, we impute Smok_Days_Since_Quitting by code (and not by the imputer), so:
- GIBBS cannot take it into account, and
- for our test Smok_Days_Since_Quitting is never missing Furthermore, as you can see in the research summary of the LEAN model, Smok_Days_Since_Quitting alone cuts significantly the gap between with and without smoking features.
MAIN RESULT - no significant change in performance ....
Example from 1 run:
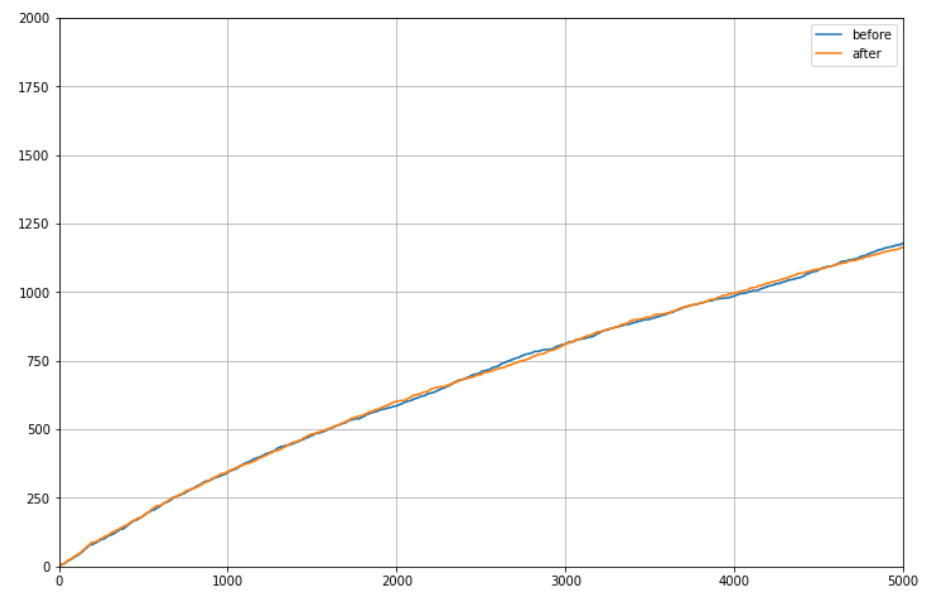
- axis x - N
- axis y- cancer detected
- parameters (most important - results are not sensitive to these parameters ...):
- sample #09
- M = 2000 (how many data completions)
- m = 20 (how many closest friends to randomly chose from for imputation)
- W - 5 for age, 3 for smoking features and BMI, 1 for labs
- Impute from future - True
- Priority for impute - pred_0 + std by GIBBS
Statistics over 10 datasets, each with 3 run (imputation has random):
 Side notes
Side notes
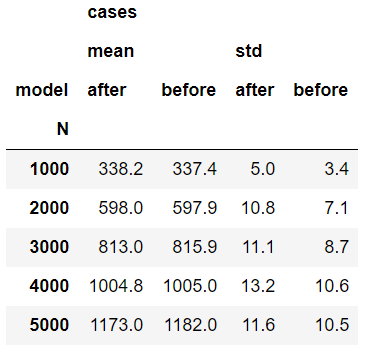
- The 'hope for success' was in the data completion procedure, as we complete missing values conditioned on the actual status (case/control). Indeed, for patients with data completion, the average score for cases increased more than for controls, but apparently not more enough. See example in the following table - change of score for M=5000
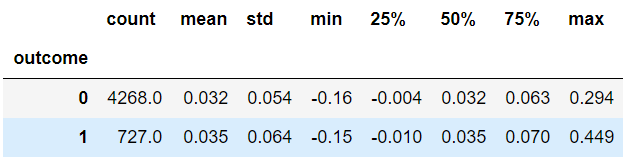
- The data completion increases the cutoff score for Top N. It makes sense, as the standard imputation impute to the mean, and we sample from the distribution, hence have more extreme results.
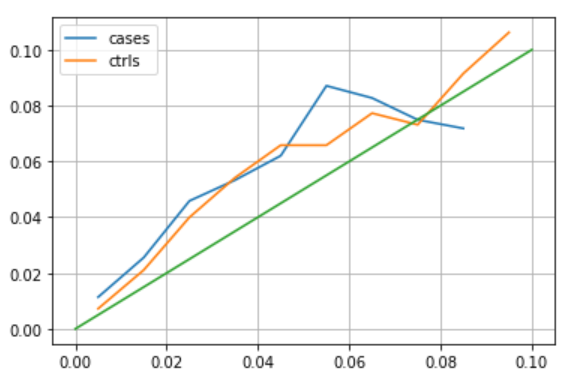
- Is the data completion by closest friends OK? Other then exploring some examples, we compare the std in score calculated in GIBBS, and the actual std we get (among patients with expected similar std). See the result in the graph - axis x is GIBBS and axis y is what we get. Trend is similar but we higher variability, could be because GIBBS calculate the std on an over fitted model (trained over all the data)