Test 05 - Compare Repository with Reference Matrix
Purpose
Compare the generated repository against a reference matrix to quantify dataset differences and identify features that drive separation. This helps estimate likely impacts on model performance and prioritise follow-up checks.
Required Inputs
From configs/env.sh:
WORK_DIR: Output folder path where compare outputs will be writtenMODEL_PATH: Path to the fitted model (the script uses${WORK_DIR}/model/model.medmdlif present)REFERENCE_MATRIX: Full path to the reference matrix (feature matrix from model training)CMP_FEATURE_RES: Comma-separated list of important features and resolutions (e.g.,Age:1,MCH.slope.win_0_1000:0.01). Used to build a restricted separation model limited to important features.
How to Run
From your TestKit folder, execute:
Outputs appear under ${WORK_DIR}/compare and ${WORK_DIR}/compare.no_overfitting.
What This Test Does
Using the TestModelExternal workflow, the test performs two comparisons:
- A full comparison using all model features (output:
${WORK_DIR}/compare). - A restricted comparison that uses only the important features derived from
CMP_FEATURE_RES(output:${WORK_DIR}/compare.no_overfitting).
For each comparison the test:
- Computes per-feature statistics (mean, std, missing counts) and statistical tests (Mann–Whitney U)
- Trains a separation/propensity model to measure how separable the new dataset is from the reference (reports AUC and related metrics)
- Produces a Shapley-style feature importance report for the separation model (
shapley_report.tsv) and per-feature plots (features_diff) - Creates HTML diagnostic reports; the script prepares a local Plotly HTML template and patches script paths so generated HTML is portable within the docs site
The compare run is guarded by timestamp checks and will skip re-running if outputs are newer than inputs, unless overridden by the test environment.
Output Location
${WORK_DIR}/compare— full comparison (all features)${WORK_DIR}/compare.no_overfitting— restricted comparison using features listed inCMP_FEATURE_RES
Key artifacts inside each directory:
compare_rep.txt— per-feature textual summary and teststest_propensity.bootstrap.pivot_txt— separation model / propensity results (AUC etc.)shapley_report.tsv— feature importance ranking for the separation modelfeatures_diff/— plots comparing distributions for top differing featuresButWhy/— HTML dashboards produced from the shapley report
How to Interpret Results
The goal is to detect distributional shifts that might affect model performance and to identify the features responsible. Use the outputs as follows:
compare_rep.txtcontains two sections: a summary block and a tabular section with per-feature stats and Mann-Whitney p-values. The tabular section is typically loaded into a DataFrame for analysis.- Mann-Whitney p-values point to distributional differences but do not capture all changes (e.g., variance differences with similar medians). Treat p-values as a signal for inspection, not automatic rejection.
test_propensity*outputs report how separable the datasets are. AUC ~ 0.5 indicates little separation (preferred). Very high AUC (e.g., ~1.0) indicates strong separation and suggests problems for matching-based performance estimation in next steps.shapley_report.tsvandfeatures_diff/plots are used to prioritise features for investigation. Always checkAgefirst as many differences can be age-proxies.
When AUC is high, prefer the restricted results in compare.no_overfitting (which isolates important features) to assess likely impact on model performance.
AUC_Meanclose to 0.5 is desirable - values near 1.0 require investigation.- Use
shapley_report.tsvto find which features drive separation. If those features are important to the model, further action is needed. - Visualize suspect features using
features_diffplots to confirm distributional shifts and check for imputation artifacts.
Suggested code to analysis compare_rep.txt
Run the following code to turn compare_rep.txt into readable dataframes.
 The second dataframe, t22, shows the same information (without range), plus Mann Whitney test result.
The second dataframe, t22, shows the same information (without range), plus Mann Whitney test result.
- _1 is for the new dataset
- _2 is for the reference
-
The Mann-Whitney U Test assesses whether two sampled groups are likely to derive from the same population, but note test limitations - if median and shape are the same for both samples, P_value would be high even for different std/scale.
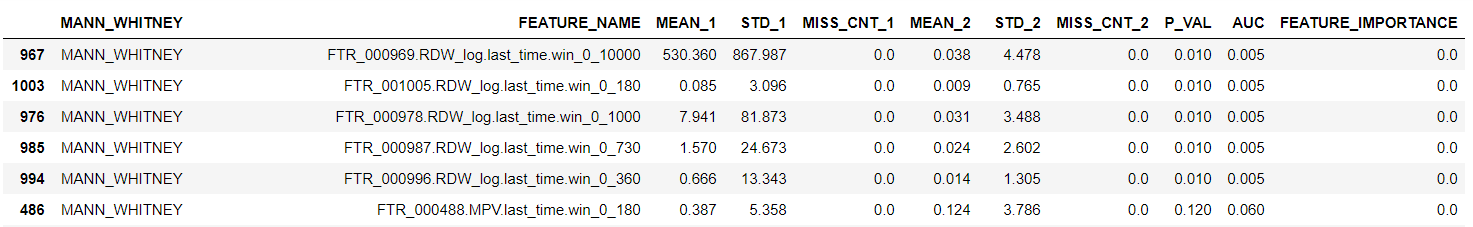 In table t22: t21 shows moments, range, and missing value counts for every feature, comparing reference to new dataset.
In table t22: t21 shows moments, range, and missing value counts for every feature, comparing reference to new dataset. -
We need to make sure we don't see low P_value for any important feature to the model, or proxy for such features, i.e.. we may list MCH.min.win_0_180 as important feature, and we don't want it or MCH.min.win_0_360 to have low P-value.
- We need to understand the reasons for the low P-value when happened, in order to better understand the new data set. For instance, in the table above, we see that in the tested dataset RDW was not given close to sample point, probably because it is not part of the panel. As RDW is not an important signal, we can ignore it.
Example
AUC_Mean is 0.98 - very high. However no significant different seen in compare_rep.txt Here is the Graph of a specific signal with big difference form shapley analysis:
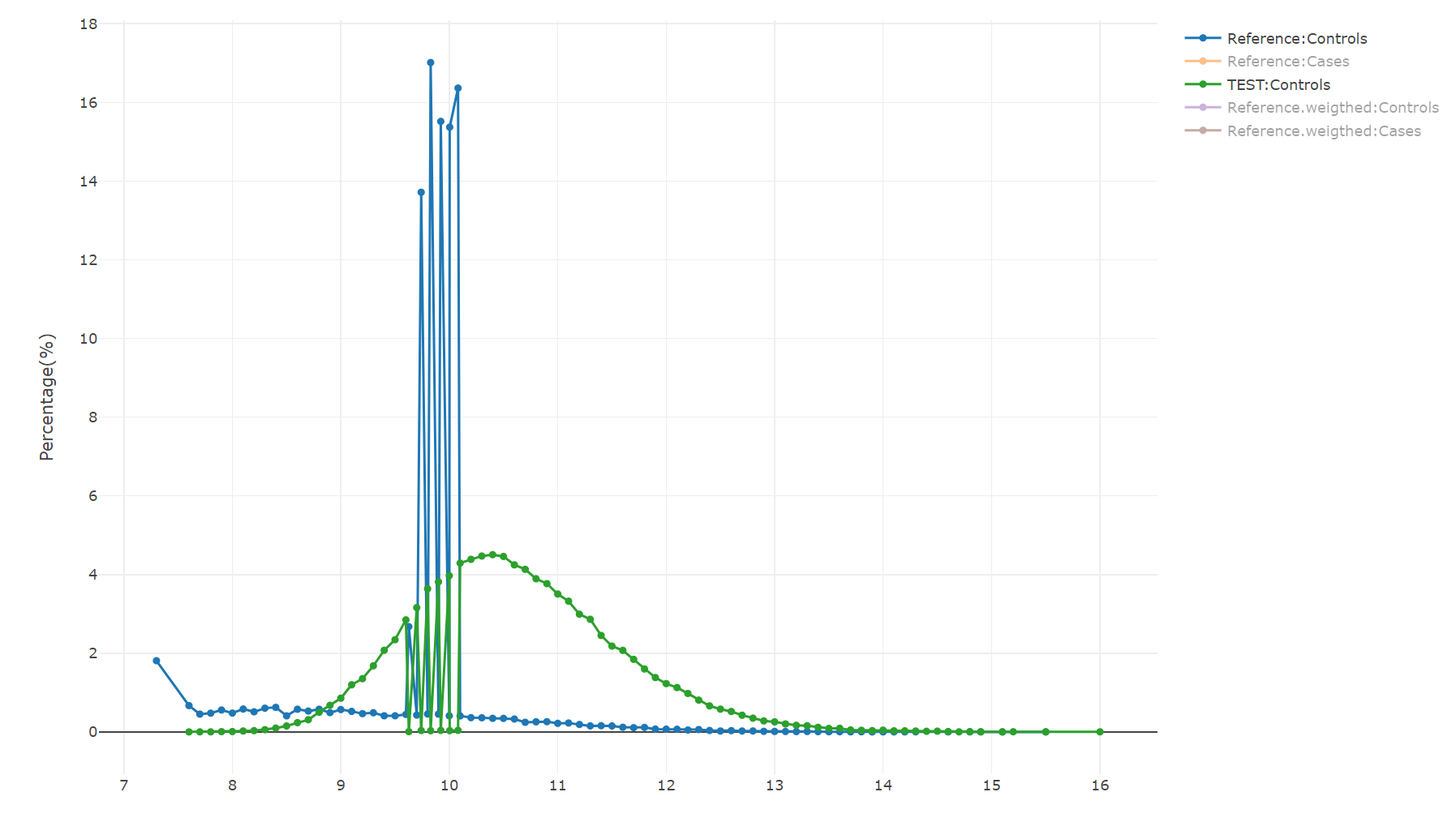 We see that the reference has several dominant values and this is due to imputations.
We see that the reference has several dominant values and this is due to imputations.
- In this case we use as reference a dataset different from the one we use for model training and feature importance.
- However, the reference dataset has many missing values for the relevant signal and imputations were generated that resulted in a binned, different resolution and distribution of the feature. It is of cource undesired situation, but not neccaraly very bad. The model was performing well in the dataset in spite of those missing values and imputations.
- Lesson learned is that we need to use as reference the original dataset (test samples only).
The reference matrix was generated again and than the AUC of this comparising dropped significantly.
Notes and Implementation Details
- A temporary Plotly-based HTML template is created at
${WORK_DIR}/tmp/plot_with_missing.htmland patched so HTML reports referencejs/plotly.jsrelative to the documentation output. CMP_FEATURE_RESis parsed into a feature list (${WORK_DIR}/tmp/feat_list) which the restricted comparison uses.- The test uses subsampling limits (
--sub_sample_test,--sub_sample_train,--sub_sample_but_why) to bound runtime and memory; these can be tuned in the script.
Troubleshooting
- If
TestModelExternalorfeature_importance_printer.pyare missing, make sure the TestKit resources are available on PATH inconfigs/env.sh. - Slow runs or memory errors: reduce subsample sizes (
MAX_SIZEin the script) or increase available memory for the Python process. - Very high AUC but no obvious per-feature differences: check whether irrelevant features produce separation (use
compare.no_overfitting) or whether imputation/binned reference values are causing artifacts. - Broken HTML reports: confirm
${WORK_DIR}/tmp/plot_with_missing.htmlexists and thatjs/plotly.jsis reachable relative to the generated report locations.
Test Results Review
Primary files to inspect after running this test:
${WORK_DIR}/compare/compare_rep.txt${WORK_DIR}/compare/test_propensity.bootstrap.pivot_txt${WORK_DIR}/compare/shapley_report.tsvand${WORK_DIR}/compare/ButWhy/${WORK_DIR}/compare/features_diff/
And for the restricted comparison:
${WORK_DIR}/compare.no_overfitting/compare_rep.txt${WORK_DIR}/compare.no_overfitting/shapley_report.tsvand.../ButWhy/
If additional examples or parsing snippets are desired (for example, the Python snippet used historically to parse compare_rep.txt), add them here. @@@[PLEASE_COMPLETE]